
![]() Spent an interesting Wednesday afternoon, last week, visiting exhibitors at the Bio-IT World Conference & Expo –several of whom won “Best of Show” Awards later that day.
Spent an interesting Wednesday afternoon, last week, visiting exhibitors at the Bio-IT World Conference & Expo –several of whom won “Best of Show” Awards later that day.
The judges, listed below, named winners in six categories: Data Integration & Management; Analysis & Data Computing; Genomic Data Services; Data Visualization & Exploration; Storage Infrastructure & Hardware; and the Judges’ Prize. Attendees also voted on the People’s Choice Award, selecting products that they believed measurably improve workflow or capacity, enabling better research.
One of my favorites was Nanome, which won best in show for Data Visualization and Exploration.
 Nanome uses virtual reality to improve the drug discovery process, according to its award application. The company offers applications for experimentation, collaborattion, and learning at the nano-scale– leveraging VR hardware such as the Oculus Rift and HTC Vive to create immersive virtual workspaces allowing users to visualize, design, and simulate molecules, proteins, and more.
Nanome uses virtual reality to improve the drug discovery process, according to its award application. The company offers applications for experimentation, collaborattion, and learning at the nano-scale– leveraging VR hardware such as the Oculus Rift and HTC Vive to create immersive virtual workspaces allowing users to visualize, design, and simulate molecules, proteins, and more.
At Nanom’se i BIO–IT World booth, Marketing Director Jarrell James handed me a pair of VR goggles and two joysticks (?) with which I could explore within a molecule–by seeming to make components larger, smaller or revolve.
A more sophisticated user might be able to:
- -Import molecular structures from a local machine or an online database such as RCSB or DrugBank.
- – Manipulate molecular structures by literally grabbing, rotating, or enlarging the area of interest with their hands.
- – Apply different representations to their selection of Atoms, Residues, Chains, or Proteins such as Stick, Wire, Ball & Stick, or Van der Waals.
- – Measure distances and angles between atoms.
- – Mutate amino acids and cycle through rotamer libraries.
- – Design small molecules by building with any element from the periodic table.
- – Minimize manipulated molecules to prevent clashes and provide a local energy minimum conformation.
- – Duplicate or Split any selected area of your structure to modify or export independently.
- – Export your molecular structures to PDB.
- – Join a virtual reality session as a guest with or without virtual reality hardware.
- – Present and collaborate in the same virtual environment with colleagues to demonstrate proposals or compare before and after results.
Nanome plans next to enter the education space. The company’s VR technology wil help high school and college students , likely already proficient in gaming technology, better understqand biologic processes, James said.
I also spent some time with the folks at Hyve…whose fake robot ( that is, a “robot inhabited” by a human) did make me curious about Hyve’s work.
As described in the company’s award submission, the company’s RADAR-base, developed in the framework of the IMI RADAR-CNS project, is an open source platform designed to securely collect, store and share readings from wearable devices and smartphone sensors to enable remote monitoring. The RADAR-base platform consists of three major categories of components:
- Data ingestion: Recognizing and registering data-sources (including smartphones and wearable devices), collecting the data via a direct Bluetooth connection or through a 3rd party API and streaming in near real time to the server (green box in the figure). Using Apache Kafka, the collected data is streamed to dedicated topics in real-time where the data is optimally schematized using Apache Avro;
- Data storage and management: Consists of two centralized storage systems behind an authorized security layer. A cold-storage based on HDFS that is scalable and fault-tolerant focusing on storing large volumes of high frequency raw-data, and a hot-storage based on MongoDB storing aggregated data to provide a near real-time overview of the raw-data. (blue box in the figure);
- Data sharing: Visualizing aggregated data in a live dashboard and exporting raw data for further analyses in various formats including AVRO, JSON and CSV (yellow box in the figure).

The platform is highly secured by a centralized management system of users and their authorities, participants, allowed devices and their specifications. RADAR-Base platform is distributed as Docker containers with associated scripts and configuration files to enable easy installation.
 In addition, I visited Sinequa, which took the prize in the Analysis & Data Computing category.
In addition, I visited Sinequa, which took the prize in the Analysis & Data Computing category.
Sinequa ES v10
sinequa.com
The Sinequa Cognitive Search and Analytics platform handles all structured and unstructured data sources and uses Natural Language Processing (NLP), statistical analysis and Machine Learning (ML) in order to create an enriched “Logical Data Warehouse” (LDW). This LDW is optimized for performance in delivering rapid responses to users’ information needs. Users can ask questions in their native language or ask that relevant information be “pushed” to them in a timely fashion when it emerges.
More than 180 connectors ready for use “out of the box” make the process of connecting multiple data sources fast and seamless. Company and industry-specific dictionaries and ontologies can be easily integrated, putting specific knowledge “under the hood” of the Sinequa platform, making it an intelligent partner for anyone in search of relevant subject information.
Other awards, as descrbed in company literature: :
Genomic Data Services
Diploid
Moon 1.0
diploid.com/moon
Moon is the first software to autonomously diagnose rare diseases from WES/WGS data. By applying AI to the domain of rare disease diagnostics, Moon brings speed and scalability to the genome interpretation process.
The software only requires the patient’s gender, age of onset and his/her symptoms – in addition to the genetic data. Moon then goes from whole genome variant data (VCF) to pinpointing the causal variant in less than 5 minutes.
The software highlights one or a few variants that could explain the patient’s phenotype. For every variant, Moon displays an extensive list of annotations that it mined from the literature, allowing geneticists to easily verify decisions from the AI algorithms. Moon’s speed does not only save a lot of time and money, it also saves lives: Moon has already proven its utility in the NICU at Rady Children’s Hospital (San Diego): https://goo.gl/7TDrQD.
Unfortunately, about 50% of rare disease patients remain undiagnosed, even after whole genome sequencing and expert interpretation. Most hospitals don’t have the resources to keep analyzing negative cases even though new correlations between genes and disorders are published every day. Moon changes all this: as the software autonomously mines the literature and analyses samples, it can reanalyze older, negative cases in the background. Only when new information that might lead to a diagnosis becomes available, the assigned geneticist is notified. That way, hospitals can frequently reanalyze thousands of cases with minimal labor, providing a perspective to undiagnosed patients.
Storage Infrastructure & Hardware
PetaGene
PetaSuite Cloud Edition – Version 1.2
petagene.com
Launching at Bio-IT World 2018, PetaSuite Cloud Edition (CE) combines two innovations: (i) the ability for a user’s software tools and pipelines to seamlessly integrate with a wide variety of cloud platforms without modification, and (ii) significantly improved, high-performance, scalable PetaSuite genomic compression technology. 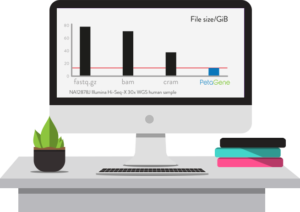
For example, users can now directly run, without modification, their custom BWA-mem, GATK, Python, Java, shell scripts, and other POSIX-based software/pipelines streaming directly to/from AWS, Google Cloud, Azure, and private cloud storage, as though they were local filestores. PetaSuite CE supports each platform’s object encryption during transfer and at rest. User applications can connect to multiple cloud platforms, buckets and regions as desired, transparently, and on demand, in user-mode, without needing to modify their pipelines, setup mounts, or have administrator privileges.
Whether running on bare-metal, in VMs, or within Docker containers, for public, private or hybrid cloud, PetaSuite CE enables organizations to unlock the power of distributed object storage seamlessly from their POSIX-compliant tools and pipelines.
PetaSuite CE is built from the ground-up for the extremely high performance streaming and random-access workloads demanded by genomics applications. The integrated, transparent PetaGene compression has been significantly improved to deliver even faster compression and greater reductions of up to 6x of both BAM and FASTQ.GZ files, enabling large costs savings in cloud storage and data transfer times. Moreover, PetaGene compression can also preserve the MD5 checksum of the original BAM or FASTQ.GZ file and not just the internal raw SAM/FASTQ data.
The Judges’ Prize went to
Linguamatics and its iScite 2.0 (iscite.com) provide a Software-as-a-Service search application that puts the power of text analytics directly into scientists’ hands, according to the company writeup.
Using Linguamatics’ Award-winning Natural Language Processing
Researchers can extract and analyze relevant data to rapidly answer business-critical questions. iScite utilizes Linguamatics’ award-winning Natural Language L(NLP) based blend of analytical methods. By understanding the semantics and structure of text, iScite handles the variety of ways people express the same information, ensuring searches are comprehensive and accurate.
Easy to use on any device
iScite’s intuitive HTML interface includes a simple search box and auto-complete suggestions. The innovative answer-routing engine lets users answer simple or complex questions using puzzle-piece building blocks – simplifying access to powerful queries that extract concepts, relationships, numerical data such as drug dosages, mutations and more.
Get answers to questions, not just documents
Data sources include Linguamatics’ cloud-hosted content. MEDLINE, Clinical Trials.gov, FDA Drug Labels, PubMed Central, and Patent Abstracts are annotated with curated terminologies for diseases, drugs, genes and organizations. Scientists can answer questions such as:
- What genes are involved in breast cancer?
- What protocol designs have been used for immuno-oncology trials?
- What are the adverse events for kinase inhibitors?
Actionable results
Results are presented in structured form, with bar chart facets for dynamic, visual results-filtering, a document viewer that highlights key terms and relationships, and relevant link-outs. Users can curate, save, and export their results.
iScite allows users across drug discovery and development to cut through the vast information landscape and discover the most valuable insights.
The People’s Choice award went to
OnRamp BioInformatics, Inc. and itsROSALIND™ platform: the first-ever genomics analysis platform specifically designed for life science researchers to analyze and interpret datasets, while freeing up more time for bioinformaticians.
 Named in honor of pioneering researcher Rosalind Franklin, who made a major contribution to the discovery of the double-helix structure of DNA with her famous photograph 51, OnRamp’s ROSALIND platform aims to simplify the practice of genomic data interpretation. According to the company’s writeup, ROSALIND puts the researcher into the driver’s seat of data analysis and democratizes bioinformatics by broadly expanding access to genomic and proteomic technologies for cancer research, precision medicine and sustainable agriculture.
Named in honor of pioneering researcher Rosalind Franklin, who made a major contribution to the discovery of the double-helix structure of DNA with her famous photograph 51, OnRamp’s ROSALIND platform aims to simplify the practice of genomic data interpretation. According to the company’s writeup, ROSALIND puts the researcher into the driver’s seat of data analysis and democratizes bioinformatics by broadly expanding access to genomic and proteomic technologies for cancer research, precision medicine and sustainable agriculture.
While many open-source tools remain the lifeline of genomic analysis, a simplified and innovative user experience for the biologist can empower them to run their own analyses, while utilizing these tools without the need for typing any command-line instructions.
ROSALIND is powered in partnership with Google Cloud and features scalable compute power and economical cloud-based storage. ROSALIND is a swarming docker-based genomic analysis solution incorporating the industry’s most trusted open-source tools and algorithms, with an angular front-end and secure RESTful API. ROSALIND is also deployable on-premise.
On Ramp technololgists believe that empowering biologists with “an intuitive and comprehensive platform” to explore their data and collaborate with colleagues and bioinformaticians, they can help accelerate their industry and the widespread adoption of genomic technologies by dramatically lowering costs, reducing complexity and, ultimately, focus more on what what to do with results, rather than on how to get to them.
In the words of Allison Profitt, BIO-IT World’s editor,” The awards program recognizes the best of the innovative product solutions for the life sciences industry on display at the conference,
“It’s always a treat to explore what’s new in our industry.
” The innovation on display by Bio-IT World exhibitors never disappoints, and we are excited to shine a spotlight on the best life sciences has to offer.”
Judges
“The Best of Show program relies on a panel of expert judges from academia and industry who screen eligible new products and hear presentations from a list of finalists on site. This year our judges considered 46 new products and viewed presentations on site from 18 finalists.”
The 2018 judging panel included Joe Cerro, BostonCIO; Chris Dwan, Bridgeplate; Richard Holland, New Forest Ventures; Eleanor Howe, Diamond Age Data Science; Phillips Kuhl, Cambridge Healthtech Institute; Steve Marshall, Marshall Data Solutions; Michael Miller, Genentech; Art Morales, Analgesic Solutions; Nanguneri Nirmala, Tufts University School of Medicine; Alexander Sherman, Massachusetts General Hospital; Subi Subramanian, Vertex Pharmaceuticals; Bill Van Etten, BioTeam; and Proffitt.
–Anita M. Harris
Anita Harris is a science writer based in Cambridge, MA.
New Cambridge Observer is a publication of the Harris Commmunications Group, also in Cambridge, ma.





